[ad_1]
The presence of Mycoplasma salivarium in the lower respiratory tract of ventilated patients with COVID-19 infection is associated with an increased chance of dying. The result was part of a molecular investigation that looked at how airway microenvironments affected severe acute respiratory syndrome (SARS-CoV-2).
Leopoldo N. Segal and his colleagues suggest that lung microbes could predict severe COVID infection and resistance to antibodies.
“These data highlight the importance of the abundance of SARS-CoV-2 in the lower respiratory tract as a predictor of mortality, and the significant contribution of the host cell transcriptome, which reflects the response of lower respiratory cells to the infection, “the researchers wrote.
The findings could help identify patients at highest risk for poor clinical outcomes and provide alternative treatments early on.
The study “Microbial signatures in the lower respiratory tract of mechanically ventilated COVID19 patients associated with a poor clinical outcome” is available as a preprint in the medRxiv* server, while the article undergoes peer review.
Classification of microbial signatures
Samples collected from the lower respiratory tract of ventilated patients for COVID-19 infection during the first wave in New York City. They collected samples from the airways of 142 patients with COVID-19 infection.
Mycoplasma salivarium in the airways is associated with worse clinical outcomes
Using metagenomics, the researchers linked the microbes living in the lung microbiome with the clinical outcomes of the patients.
The results showed that having high amounts of Mycoplasma salivarium was associated with a higher SARS-CoV-2 viral load. Furthermore, a limited immunoglobulin response in the lower respiratory tract was correlated with an increased risk of mortality.
“The data presented here using direct quantitative methods (RT-PCR) and an undirected semi-quantitative approach (metatranscriptome sequencing) support the hypothesis that lower respiratory tract SARS-CoV-2 viral load plays a role. critical in clinical progression of critically ill patients with COVID-19, “the researchers wrote.
There is no evidence of worse clinical outcomes from coinfection with respiratory pathogens
Although the majority of patients received antibiotics and broad-spectrum antifungals, there was no evidence of worsening effects of co-infection with bacterial, viral, and fungal respiratory pathogens.
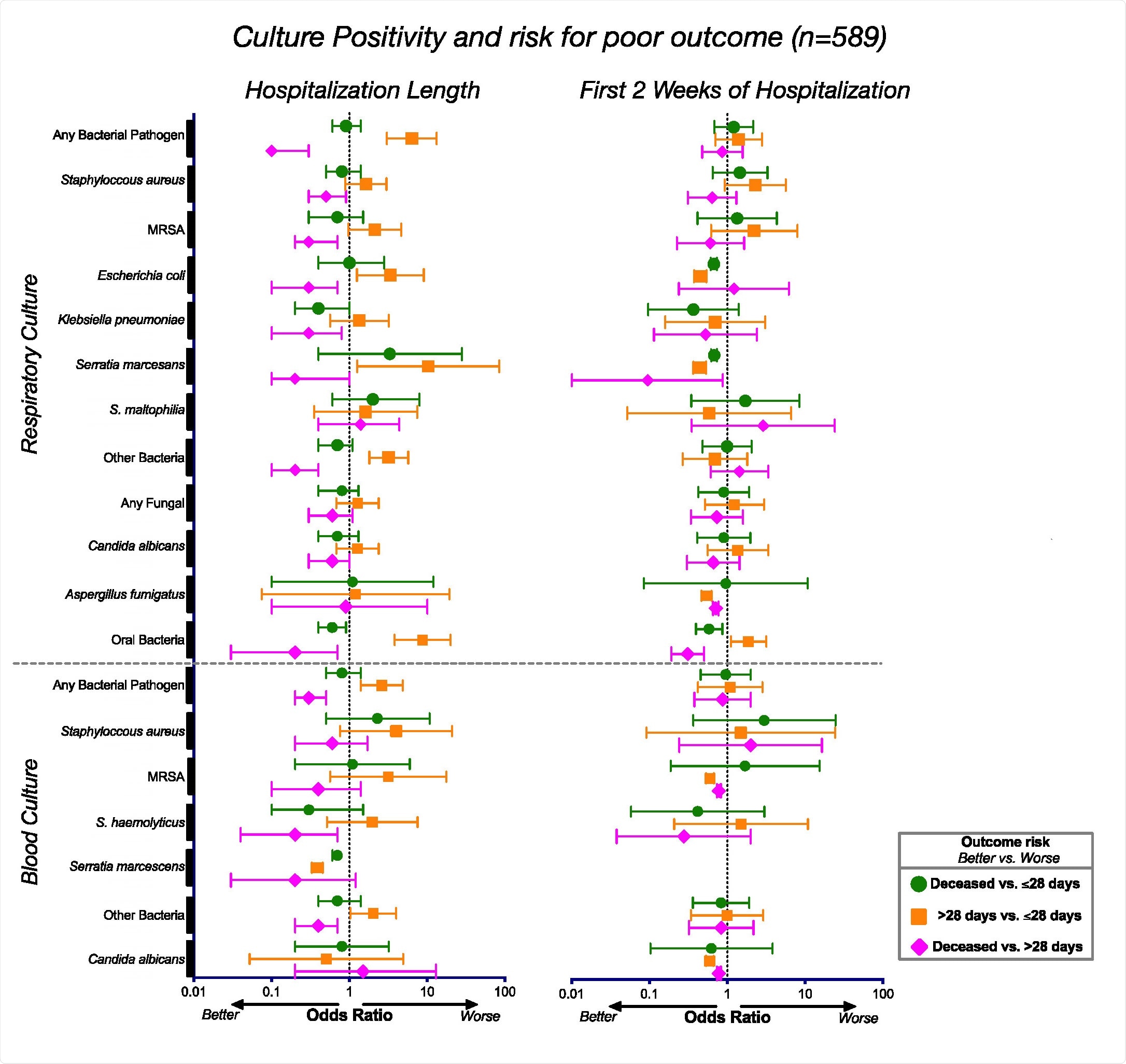
To look at the risk of mortality from COVID-19 infection, the team analyzed laboratory cultures of 589 patients hospitalized for respiratory failure due to severe COVID-19 infection.
The results showed that patients with poor clinical outcomes did not succumb to other respiratory infections. There was also no relationship between positive microbial cultures and mortality in severe COVID-19 infections.

Associations between positive culture and clinical result. Odds ratios and corresponding 95% confidence intervals for the positive culture rates for the entire cohort (n = 589) during the duration of their hospitalization (left) and during the first 2 weeks of hospitalization (right).
Bacterial changes observed in patients ventilated for more than 28 days
Beyond microbes, the lower respiratory tract showed a high presence of SARS-CoV-2, which was associated with death. A small sample of patients had influenza A or B viruses, suggesting that influenza is unlikely to occur simultaneously with coronavirus infection.
When looking at bacteria in the lower respiratory tract, the metatranscriptome data found phages actively present. The researchers suggest that this could be evidence that alterations in the bacterial microbiome could be occurring in patients with severe COVID-19 infection.
Changes were observed in Staphylococcus phages, and Mycoplasma salivarium it was actively present in patients who required ventilation for more than 28 days and in patients who died compared to patients who received ventilation for less than 28 days.
Microbial impact on the immune response
Patients with poor clinical outcomes expressed pathways that activated genes related to degradation, transport, in addition to expressing antimicrobial resistance and signaling genes.
The researchers write:
“These differences may indicate important functional differences that lead to a different metabolic environment in the lower respiratory tract that could affect the host’s immune response. It could also be representative of differences in microbial pressure in patients with higher viral loads and different environments. inflammatory “.
There was also a positive regulation in the signaling pathways of Sirtuin and Ferroptosis in the most severe cases of COVID-19. This was consistent with inactivated immune response characteristics, including phagocytes, neutrophils, granulocytes, and leukocytes. A down regulation of immunoglobulin expression levels and mitochondrial dysfunction were also observed.
Based on the data, the team suggests that the lungs of critically ill patients requiring ventilation due to COVID-19 infection express a state of imbalance rather than elevated inflammation. Doing so appears to predict a worsening of the prognosis.
A further analysis found differences associated with survival in responses to interferon. Activation of type I interferon was a predictive factor for increased mortality.
“While more longitudinal data will be needed to clarify the role of interferon signaling in disease, the data presented here suggest that the combination of host and microbial signatures could help understand the increased risk of mortality in COVID-19 patients. critically ill “.
*Important news
medRxiv publishes preliminary scientific reports that are not peer-reviewed and therefore should not be considered conclusive, guide clinical practice / health-related behavior, or be treated as established information.