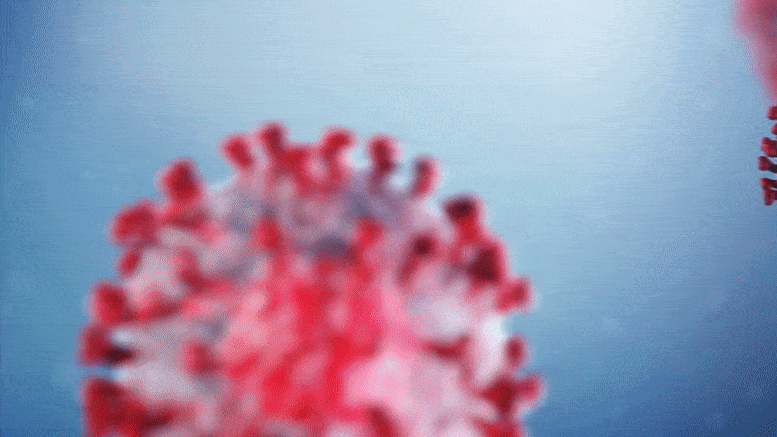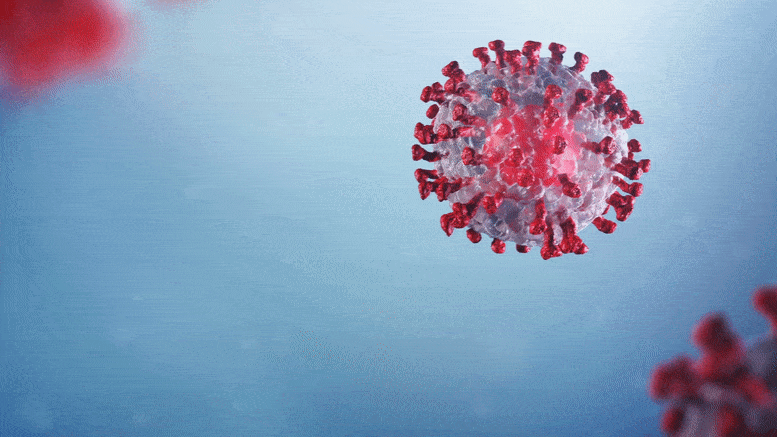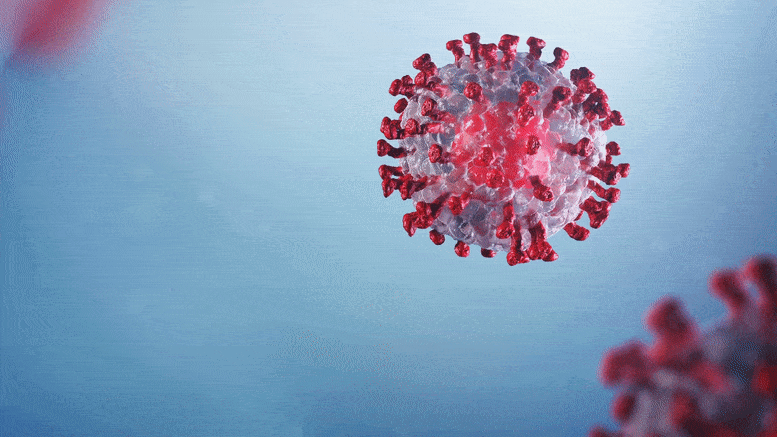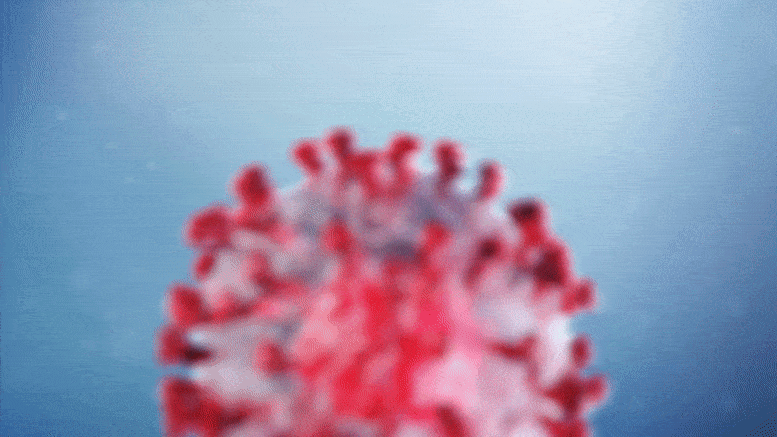

The discovery lays the foundation for designing new antiviral drugs.
With an alarm code, we can enter a building without the bells ringing. It turns out that the SARS 2 coronavirus (SARS-CoV-2) has the same advantage when entering cells. She has the code to dance the waltz.
Today (July 24, 2020), in Nature’s Communications, researchers at the University of Texas Health Sciences Center at San Antonio (UT Health San Antonio) reported how the coronavirus achieves this.

Yogesh K. Gupta, PhD, and colleagues at UT Health San Antonio discovered the mechanism by which the new coronavirus can enter cells without encountering resistance in the immune system. Credit: UT Health San Antonio
The scientists worked out the structure of an enzyme called nsp16, which the virus produces and then uses to modify its messenger. RNA cap, said Yogesh Gupta, PhD, lead author of the Joe R. and Teresa Lozano Long School of Medicine study at UT Health San Antonio.
“It is camouflage,” said Dr. Gupta. “Due to the modifications, which trick the cell, the resulting viral messenger RNA is now considered part of the cell’s own code and not foreign.”
Deciphering the 3D structure of nsp16 paves the way for the rational design of antiviral drugs for COVID-19 and other emerging coronavirus infections, Dr. Gupta said. The drugs, new small molecules, would inhibit nsp16 from making the modifications. The immune system would launch into the invading virus, recognizing it as foreign.
“Yogesh’s work discovered the 3D structure of a key COVID-19 virus enzyme required for its replication and found a pocket in it that can be targeted to inhibit that enzyme. This is a critical advance in our understanding of the virus, “said study co-author Robert Hromas, MD, professor and dean of the Long School of Medicine.
Dr. Gupta is an assistant professor in the Department of Biochemistry and Structural Biology at UT Health San Antonio and is a member of the university’s Greehey Childhood Cancer Research Institute.
In simple terms, messenger RNA can be described as a genetic code generator for protein producing work sites.
###
Reference: “Structural basis of SARS-CoV-2 RNA boundary modification” by Thiruselvam Viswanathan, Shailee Arya, Siu-Hong Chan, Shan Qi, Nan Dai, Anurag Misra, Jun-Gyu Park, Fatai Oladunni, Dmytro Kovalskyy , Robert A Hromas, Luis Martinez-Sobrido and Yogesh K. Gupta, July 24, 2020, Nature Communications.
DOI: 10.1038 / s41467-020-17496-8
The laboratory of lead author, Yogesh Gupta, PhD, is supported by funds from the Max and Minnie Tomerlin Voelcker Foundation, the San Antonio Area Foundation, the University of Texas System, UT Health San Antonio, the Institute of Greehey Children’s Cancer Research from UT Health San Antonio, and the Texas Cancer Research and Prevention Institute (CPRIT).