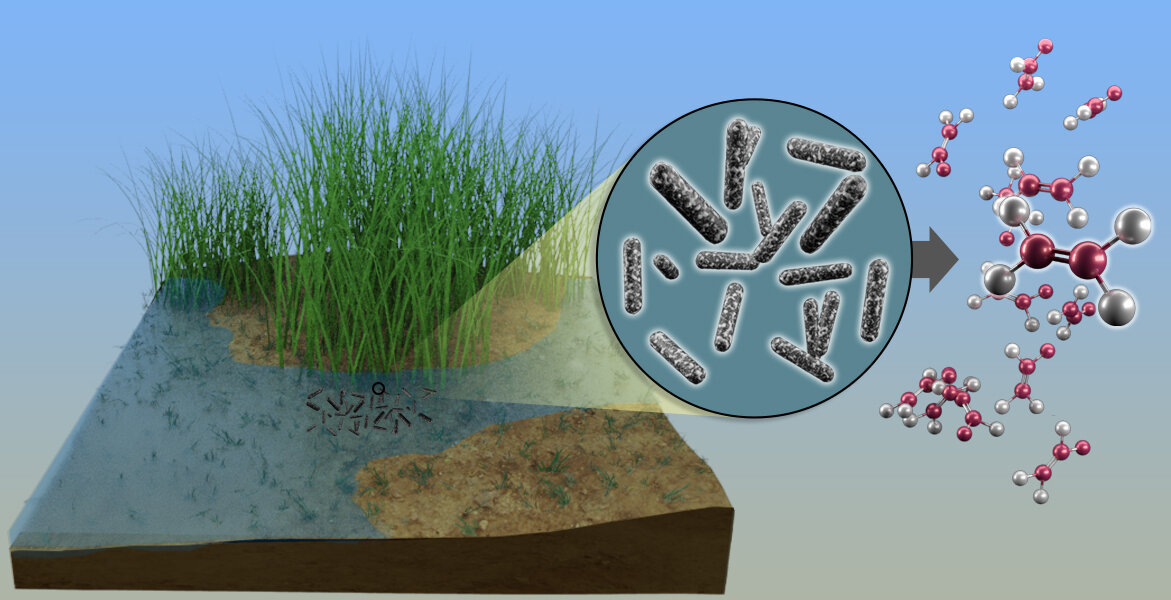

Scientists have discovered how microbes in waterlogged soils produce high levels of ethylene, which can negatively affect agricultural crops and bioenergy feedstocks such as switchgrass. This new knowledge can be used to develop treatments for healthier crops. Credit: Andy Sproles / ORNL, US Department of Energy
Scientists from the Oak Ridge National Laboratory of the Department of Energy and Ohio State University have discovered a new microbial pathway that produces ethylene, and offers a potential pathway for bioproduction of a common component of plastics, adhesives, refrigerants and other everyday products.
The discovery, published in Science, sheds light on a long-standing mystery about how ethylene is produced in anaerobic, or oxygen-decomposed, soils and points to potential pathways to prevent crop damage from high levels of ethylene. The study also outlines a previously unknown way in which bacteria generate methane, a powerful greenhouse gas.
The research team found that ethylene and methane are by-products of a bacterial process that produces methionine, an amino acid needed to build egg whites. When their environment is anaerobic and low in sulfur, bacteria are forced to remove sulfur from cellular waste products, activating this new pathway.
“For about a decade, researchers have been investigating the biological production of ethylene by another mechanism that occurs in oxygen environments,” said Ohio state research scientist Justin North. “There is a technical barrier to scaling up that process, because ethylene and oxygen mixed on an industrial scale can be explosive. This new anaerobic trajectory removes that barrier, but there is still work to be done to scale it up. . “
The research began in the state of Ohio where Robert Tabita is conducting an ongoing study on carbon fixation and nitrogen and sulfur metabolism in photosynthetic bacteria. As part of Tabita’s team, North decided to measure the gases consumed and emitted by Rhodospirillum rubrum and other microbes in the same family when they became hungry for sulfur. He was surprised to find ethylene.

ORNL’s Bob Hettich used a special mass spectrometry technique to characterize the proteomes of microbial systems. Credit: Carlos Jones / ORNL, US Department of Energy
“We know that these bacteria produce hydrogen and consume carbon dioxide,” North said. “But, look, they made a lot of ethylene gas. And we thought, well, that’s weird.”
North and his colleagues from Ohio State studied this new metabolic process with radioactive compounds to track the precursors and the production of methionine and ethylene in microbes. But another type of analytical biotechnology was needed to create the critical link between the pathway and the proteins called enzymes that drive it.
Tabita reached out to Bob Hettich, who leads the Biological Mass Spectrometry Group at ORNL, for a comparative analysis of the collection of proteins, called proteomes, present in these photosynthetic bacteria under two different scenarios: low sulfur, ethylene producing conditions and high-sulfur, non-ethylene producing conditions. The Hettich group has developed an advanced approach for characterizing the proteomes of microbial systems using mass spectrometry, a technique that accurately measures the masses and fragmentation pathways of different molecules and provides details on structure and composition. Hettich and Weili Xiong, an ORDL postdoctoral researcher, identified thousands of proteins from the low and high sulfur systems and analyzed their comparative abundance to identify a handful of eggs for further characterization.
“We found striking differences,” Hettich said. The data showed a family of nitrogenase-like proteins that were nearly 50-fold abundant in the low-sulfur, ethylene-producing samples. Some iron and sulfur-related egg whites also increased in abundance when sulfur was scarce, pointing to a possible new pathway for sulfur metabolism.
These data were surprising because nitrogenase-like egg proteins are grouped into gene tags with nitrogenases that have similar DNA sequences and are known to convert atmospheric nitrogen to ammonia. This process for nitrogen management is essential for life on earth and has been extensively researched. Given their name, these nitrogenase-like proteins are not the ones that scientists would have thought play a role in sulfur metabolism.

Weili Xiong collaborated on mass spectrometry research while at ORNL as a postdoctoral associate. Credit: Carlos Jones / ORNL, US Department of Energy
“Sometimes naming or annotating a gene as a gene family can be misleading,” Hettich said. “The name suggests a primary function. In fact, the gene may have a secondary function, a nightmare so to speak, or it may actually do something completely different.”
“But the data is the data,” he continued. “If you perform the measurements well and in an agnostic way where you do not know the answer a priori, then the data will open the real connections. “
With this crucial proteomic data, Ohio State researchers and colleagues at Colorado State University and Pacific Northwest National Laboratory conducted a series of experiments that manipulated the bacterial genome to incorporate or remove the Rru_A0793-Rru_A0796 gene cluster. The removal and replacement of genes switched on and off ethylene production as a switch, confirming that the genes and the resulting enzyme they encode are essential for this metabolic pathway.
The nitrogenase-like enzymes cleave carbon-sulfur bonds to reduce 2- (methylthio) ethanol in a precursor to methionine production. This pathway produces ethylene as a by-product. The research team found that when the source of sulfur is converted into dimethyl sulfide, the most abundant volatile organic sulfur compound, bacteria use it in their methionine pathway and produce methane as a by-product.
In addition to being a potential biological agent for the production of ethylene for use in plastics and other industrial products, these findings may inform treatments for crops in water-dried, anaerobic soils to prevent damage by an excess of ethylene. In the right amounts, ethylene is an important plant hormone that helps plants grow, develop leaves and roots and ripen fruits. This study raises a host of new scientific questions, including whether this pathway is involved in interactions between plants and microbes.
“It is very exciting that this discovery is leading to new lines of research that may actually have some substantial benefits for agricultural and other crops as well,” North said.
Researchers discover new ethylene signaling mechanism in rice roots
“In nitrogenase-like enzyme system, methionine, ethylene, and methane catalyze biogenesis” Science (2020). science.sciencemag.org/cgi/doi… 1126 / science.abb6310
Delivered by Oak Ridge National Laboratory
Quote: Sulfur-scavenging bacteria may be the key to making common component in plastic (2020, August 27) Retrieved August 28, 2020 from https://phys.org/news/2020-08-sulfur-scavenging-bacteria-key- common-component .html
This document is subject to copyright. Except for any fair trade for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.