 Portrait of Red Chalk in the Man (1512). “/>
Portrait of Red Chalk in the Man (1512). “/>Guadalupe Pier et al.
Microbiomes are all the rage in science, even in art conservation, where studying the microbial species that accumulates on works of art can lead to new ways to slow down the deterioration of invaluable aging artwork, as well as potential unmasked forgeries. For example, scientists have analyzed microorganisms found on seven drawings of Leonardo da Vinci, according to a recent paper published in the journal Microbiology. And back in March, j. Scientists at the Craig Venter Institute (JCVI) collected and analyzed swabs from centuries-old art in a private collection in Florence, Italy, and published their findings in the Journal of Microbial Ecology.
The researchers behind the previous March paper were JCVI geneticists who collaborated with the Leonardo da Vinci DNA project in France. Work built on prior study to find microbial signatures and possible geographical patterns in hair collected from people in the Columbia District and San Diego, California. They concluded from that analysis that microbes could be a useful geographical signature.
For the March study, JCVI geneticists took microorganisms from Renaissance-style fragments and confirmed the presence of so-called “oxidase positive” microorganisms on painted wood and canvas surfaces. These microorganisms attack the compounds found in paints, glues and cellulose (found in paper, canvas and wood), in turn producing water or hydrogen peroxide as byproducts.
“Such byproducts affect the presence of mold and the overall rate of deterioration,” the authors noted in their paper. “Although previous studies have attempted to characterize the microbial composition associated with artwork decay, our results summarize the first large-scale genomics-based study to understand the microbial communities associated with aging artwork.”
As an added bonus, they found that they could distinguish between microbial populations on different types of materials. In particular, the art of stone and marble stimulated a more diverse population than paintings, possibly with the porous nature of the stone and the potential moisture and nutrients found in the marble, along with the potential for biofilm, ”he wrote. To provide more insignificant nutrients.
The authors acknowledged the small sample size, but nevertheless concluded that microbial signatures could be used to differentiate artwork depending on the material. As always, more research is needed. “Of particular interest will be the presence and activity of oil-reducing enzymes,” the authors wrote. “Such approaches will give a full understanding of which organisms (s) are responsible for the rapid decay of artwork when potentially this information is used to target this organism.”
Swabbing Renaissance Art
-
J. Collage of various art works sampled for the March Paper by geneticists at the Craig Venter Institute. Circles indicate areas swabbed on each sample artwork
JCVI
-
Manoloto G. to collect microbes from centuries-old, Renaissance-style art at a private collector’s house in Florence, Italy. Toralba and his colleagues used small, dry polyester swabs.
Jesse us subel
Guadalupe Pierre and his team collaborated with the Instituto Central de la Patologa Digli Archive e del Libro (ICPL) at the University of Natural Resources and Life Sciences in Austria for microbiome analysis of Leonardo da Vinci drawings. Last year, Pierre Et al. Relied on microbiome analysis to study the storage status of the three statues obtained from smugglers as well as to direct their possible geographical origins. Earlier this year, they analyzed the microbiome of 1,000-year-old parchment, from which they were able to cut animals whose skins are used to make parchment.
For this latest paper, Pierre’s team identified a third-generation sequencing method called a nanopore, using protein nanopores embedded in a polymer membrane for sequencing. It comes with a portable, pocket-sized sensing device, the Minion, making it ideal for studying cultural heritage. For Leonardo drawing, Pierre Et al. Combined nanopore sequencing with the whole genome-amplification protocol.
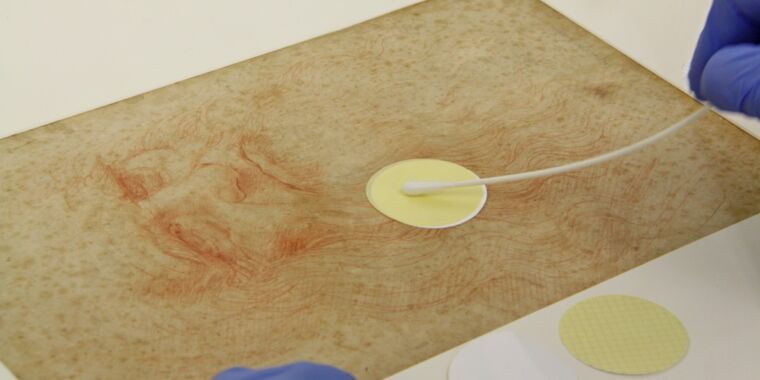
ICPL conservators used a delicate, noninvasive microspiration (i.e. filtering suction) sampling method to collect dust particles, microbial cells and other debris from small surface areas on both. Recto And o Latu Each picture. The Austrian team then dug up, expanded and sequenced the DNA. To analyze all the micro-objects collected from the drawings, they used ical pteroscopic microscopy for image features and scanning electron microscopy (SEM) of interest in all seven drawings.
A “biological genealogy”
-
Leonardo da Vinci’s drawings have been analyzed in a study by Austrian and Italian scientists.
Guadalupe Pier et al.
-
Sampling the microbiome of Leonardo da Vinci Figura Iginiochiaata from Un of Studio di Peneggio (Ca. 1475)
Guadalupe Pier et al.
-
Leonardo da Vinci Yumo della Bitta.
Guadalupe Pier et al.
-
Insect droppings on the fibers of the L4 drawing appear as wax-brown introstations (Studio del Gambe Anterori di un Cavallo).
Guadalupe Pier et al.
-
SEM images of the membrane surface used for L3 surface sampling (Nudi di la la batgalia di angiri).
Guadalupe Pier et al.
Each drawing had its own unique microbiome – an “independent molecular profile or biological genealogy.” But Pierre Et al. Contrary to the widespread belief that fungi would become more prevalent due to the high probability of colonization on paper-based works, overall, I was surprised to learn that bacteria dominated fungi in the microbiomes of drawings. Researchers have found no visible biodegradation on the drawing other than foxing stains.
Most of these bacteria are commonly found in human microbes, indicating that they found their way to drawings when handled during a restoration – although one can only guess whether the artist himself came. (The authors note that bacteria in the dust can stay in “suspension” for a long time.) Other bacteria were typical of insect microbiomes and were introduced long ago by flies that accumulate excrement on drawings. Those droppings have been shown as wax brown introstation in the fibers under imaging analysis.
The Strian / Italian team was unable to conclusively conclude whether any microbes were contaminated by Leonardo’s time. It seems that the human microbial elements are due to the recent restoration work. This could most likely be the case for the drawing designated L4 (Studio del Gambe Anterori di un Cavallo) In particular, which showed the least contamination from biodiversity and human DNA. The authors further speculate that the recipe Leonardo uses – “an initial layer of powdered calcined chicken bones, white lead, indigo … mixed with animal gelatin,” interferes with the defense of L4’s microbiome, so only recent DNA remains. .
But Pierre insists that being able to track this type of data is invaluable. “The sensitivity of the Nanopore sequencing method provides an excellent tool for monitoring art objects. It allows the evaluation of microbiomes and the visualization of its variations due to harmful conditions.” “This can be used as a bio-archive of the history of objects, providing a kind of fingerprint for current and future comparisons.”
DOI: Microbial Ecology, 2020. 10.1073 / pnas.1802831115
DOI: Frontiers in Microbiology, 2020. 10.1007 / s00248-020-01504-x (About DOI).