[ad_1]
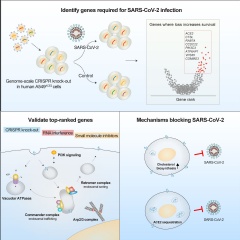
To identify potential new therapeutic targets for SARS-CoV-2, a team of scientists from the New York Genome Center, New York University, and the Icahn School of Medicine at Mount Sinai, performed a scale loss-of-function CRISPR. of the genome screened to systematically remove all genes from the human genome. The team examined which genetic modifications made human lung cells more resistant to SARS-CoV-2 infection. Their findings revealed individual genes and regulatory networks of genes in the human genome that are required by SARS-CoV-2 and that confer resistance to viral infection when deleted. The collaborative study described a wide range of genes that had not previously been considered therapeutic targets for SARS-CoV-2. Their study was published online by Cell October 24.
In order to better understand the complex relationships between the genetic dependencies of the host and the virus, the team used a wide range of analytical and experimental methods to validate their results. This integrative approach included genome editing, single cell sequencing, confocal imaging, and computational analyzes of gene expression and proteomic data sets. The researchers found that these new genetic targets, when inhibited through the use of small molecules (drugs), significantly reduce viral load and, with some drugs, up to 1,000 times. Their findings offer insights on new therapies that may be effective in treating COVID-19 and reveal the underlying molecular targets of those therapies.
?? Seeing the tragic impact of COVID-19 here in New York and around the world, we felt we could use the high-throughput CRISPR gene editing tools that we have applied to other diseases to understand what the key human genes are required by SARS-CoV-2 virus, ?? said study co-lead author Dr. Neville Sanjana, a senior faculty member at the New York Genome Center, an assistant professor of biology at New York University, and an assistant professor of neuroscience and physiology at the Grossman School of Medicine of the University of New York. Previously, Dr. Sanjana has applied genome-wide CRISPR screens to identify the genetic drivers of various diseases, including drug resistance in melanoma, immunotherapy failure, lung cancer metastasis, innate immunity, innate metabolic disorders and muscular dystrophy.
For this project, genome editing was only half of the equation. “Previously, we developed a series of human cell models for coronavirus infection in our work to understand immune responses to the virus. It was great to team up with Neville’s group to comprehensively understand and profile host genes from a new angle. said co-lead author Dr. Benjamin tenOever, Fishberg Professor of Medicine, Icahn Scholar and Professor of Microbiology, Icahn College of Medicine at Mount Sinai.
Gene clusters lead the way
The team found that the highest ranked genes ?? those whose loss substantially reduces viral infection? clustered into a handful of protein complexes, including vacuolar ATPases, Retromer, Commander, Arp2 / 3, and PI3K. Many of these protein complexes are involved in the trafficking of proteins to and from the cell membrane.
?? We were very happy to see multiple genes within the same family as top ranked hits on our whole genome screen. This gave us a high degree of confidence that these protein families were crucial to the life cycle of the virus, either to enter human cells or to successful viral replication. said Dr. Zharko Daniloski, a postdoctoral fellow at the Sanjana Laboratory and co-first author of the study.
While the researchers performed CRISPR screening using human lung cells, the team also explored whether the expression of the required host genes was lung-specific or more broadly expressed. Among the highest ranked genes, only ACE2, the receptor known to be responsible for the binding of the SARS-CoV-2 Spike viral protein, showed tissue-specific expression, and the rest of the major genes were widely expressed in many tissues, suggesting that these mechanisms may function regardless of cell or tissue type. Using proteomic data, they found that several of the best-ranked host genes interact directly with proteins from the virus itself, highlighting their central role in the viral life cycle. The team also analyzed common host genes required for other viral pathogens, such as Zika or pandemic H1N1 influenza.
Mechanistic insights: cholesterol and viral receptors
After completing the primary screening, the group of researchers used several different techniques to validate the role of many of the highest ranked genes in viral infection. Using human cell lines derived from the lung and other organs susceptible to SARS-CoV-2 infection, they measured viral infection after deletion of the gene by CRISPR, deletion of the gene through RNA interference, or drug inhibition. After validating that these manipulations reduced viral infection, they then sought to understand the mechanisms by which the loss of these genes blocks coronavirus infection.
Using a recently developed technology that combines large-scale CRISPR editing with single-cell RNA sequencing (ECCITE-seq), the team identified that the loss of several high-ranking genes results in upregulation of cholesterol biosynthetic pathways. and an increase in cellular cholesterol. . Using this information, they studied the effects of amlodipine, a drug that alters cholesterol levels.
?? We found that amlodipine, a calcium channel antagonist, upregulates cellular cholesterol levels and blocks SARS-CoV-2 infection. Since recent clinical studies have also suggested that patients taking calcium channel blockers have a reduced fatality rate from COVID-19, an important research direction going forward will be to further illuminate the relationship between the synthetic pathways. cholesterol and SARS-CoV-2, ?? said Dr. Tristan Jordan, a postdoctoral fellow at the tenOever Lab and co-first author of the study.
Building on previous work on mutations in the Spike protein and viral entry through the ACE2 receptor, the research team also asked whether the loss of some genes could confer resistance to the coronavirus by reducing ACE2 levels. They identified a particular gene, RAB7A, which has a great impact on the traffic of ACE2 to the cell membrane. Using a combination of flow cytometry and confocal microscopy, the team showed that the loss of RAB7A prevents viral entry by sequestering ACE2 receptors within cells.
“Current treatments for SARS-CoV-2 infection target the virus itself, but this study offers a better understanding of how host genes influence viral entry and will allow new avenues for therapeutic discovery and hopefully accelerate the recovery of susceptible populations “. Dr. Sanjana said.
# # #
About the New York Genome Center
The New York Genome Center (NYGC) is an independent, non-profit academic research institution that serves as a multi-agency center for genomics research. Leveraging our strengths in whole genome sequencing and genomic analysis, our mission is to advance genomic science and its application to drive new biomedical discoveries. NYGC’s focus areas include the development of experimental and computational genomic methods and disease-focused research to better understand the genetic basis of cancer, neurodegenerative diseases and neuropsychiatric diseases. In 2020, the NYGC has also directed its expertise to COVID-19 investigation.
NYGC draws on experience and draws on the combined strengths of our faculty, staff scientists, member institutions, scientific task forces, affiliate members, and industry partners to advance genomic discovery. Central to our scientific mission is an outstanding faculty who run independent research laboratories based at the NYGC and maintain joint appointments with one of our member institutions.
The institutional founding members of the NYGC are: Cold Spring Harbor Laboratory, Columbia University, Albert Einstein College of Medicine, The Jackson Laboratory, Memorial Sloan Kettering Cancer Center, Icahn School of Medicine at Mount Sinai, NewYork-Presbyterian Hospital, New York University, Northwell Health, The Rockefeller University, Stony Brook University, and Weill Cornell Medicine. Institutional Associate Members are: American Museum of Natural History, Georgetown Lombardi Comprehensive Cancer Center, Hackensack Meridian Health, Hospital for Special Surgery, The New York Stem Cell Foundation, Princeton University, Roswell Park Cancer Institute, and Rutgers Cancer Institute of New Jersey.
For more information about NYGC, visit: http://www.nygenome.org.
This news content was configured by the WebWire editorial staff. Linking is allowed.
Press release distribution and press release distribution services provided by WebWire.
[ad_2]