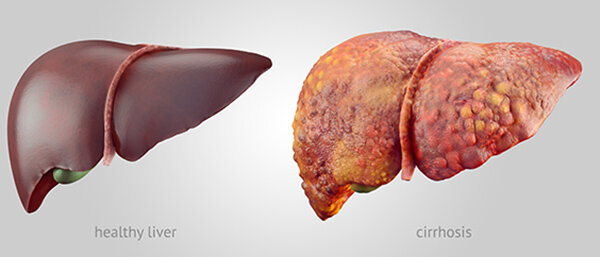

The illustration shows a healthy liver (left) and a liver with cirrhosis. Credit: National Institute of Diabetes and Digestive and Kidney Diseases.
Non-alcoholic fatty liver disease (NAFLD) is the leading cause of chronic liver disease worldwide, affecting a quarter of the world’s population. It is a progressive condition that, in the worst case, can cause cirrhosis, liver cancer, liver failure, and death.
In a new article published online June 30, 2020 in Cellular metabolism, a team of scientists, led by researchers at the University of California, San Diego School of Medicine, reports that the stool microbiomes (the collection of microorganisms found in fecal matter and the gastrointestinal tract) of patients with NAFLDs are distinct enough to be used to accurately predict which people with NAFLD are most at risk for cirrhosis: late-stage irreversible scarring of the liver that often requires eventual organ transplantation.
“The findings represent the possibility of creating an accurate, non-invasive, microbiome-based stool test to identify patients at increased risk for cirrhosis,” said lead author Rohit Loomba, MD, professor of medicine in the Division of Gastroenterology. from the University of California at San Diego. Faculty of Medicine and director of its NAFLD Research Center. “Such a diagnostic tool is urgently needed.”
Loomba said that a novel aspect of the study is the external validation of the gut microbiome cirrhosis signatures in participating cohorts from China and Italy. “This is one of the first studies to show such robust external validation of a gut microbiome-based signature across geographically distinct ethnicities and cohorts.
The work is based on research previously published in 2017 and 2019 by Loomba and colleagues.
A link between NAFLD and the gut microbiome is well documented, but details were sparse and it is not clear that discrete metagenomics and metabolomics signatures can be used to detect and predict cirrhosis. In the latest study, researchers compared the stool microbiomes of 163 participants spanning patients with NAFLD cirrhosis, their first-degree relatives, and control patients without NAFLD.
By combining the metagenomic signatures with the ages of the participants and the levels of serum albumin (an abundant blood protein produced in the liver), the scientists were able to accurately distinguish cirrhosis in participants who differ by cause of disease and geography.
The next step, Loomba said, is to establish the causality of these intestinal microbial species or their metabolites to cause cirrhosis, and whether this test can be used and expanded for clinical use.
Fatty liver disease is undiagnosed in the U.S.
A signature derived from universal gut microbiome predicts cirrhosis, Cellular metabolism (2020). DOI: 10.1016 / j.cmet.2020.06.005, www.cell.com/cell-metabolism/f… 1550-4131 (20) 30306-5
Provided by the University of California – San Diego
Citation: The universal signature derived from the intestinal microbiome predicts cirrhosis (2020, June 30) retrieved on July 1, 2020 from https://medicalxpress.com/news/2020-06-universal-gut-microbiome-derived-signature- cirrhosis.html
This document is subject to copyright. Other than fair dealing for private research or study purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.